Introduction
The human gut hosts the largest scale of symbiotic microorganisms in the body, among which the bacterial population reaches up to one hundred billion per milliliter in the colon (Sender et al., 2016). The intestine contains bacteria such as Streptococcus, Lactobacillus, Bacteroides, Bifidobacterium, and Akkermansia, with the number of species amounting to 1,952 uncultured bacteria and 553 known colonizers (Almeida et al., 2019).
Certain microbes isolated from the intestines of healthy individuals are being developed into pharmaceuticals, targeting conditions such as gastrointestinal disorders, dental disorders, and conditions in infants (Cordaillat-Simmons et al., 2020). Traditional therapeutic approaches have encountered challenges, including mounting antibiotic and chemotherapy resistance, drug non-responsiveness, and limited specificity. In contrast, microbiome-based treatments offer promising solutions, as they surmount these limitations (Yadav and Chauhan, 2022).
Research results are being published indicating that the balance of intestinal microbial communities is closely interconnected with diseases. The composition of the gut microbiome is associated with the potential development of various conditions, including cancer (Tilg et al., 2018), autoimmune diseases (De Luca and Shoenfeld, 2019), metabolic syndrome (Fan and Pedersen, 2021), inflammatory bowel disease (Glassner et al., 2020), and neurodegenerative diseases (Zhu et al., 2021), by influencing gut permeability and mucosal immunity (Zheng et al., 2020). Next-generation sequencing has been utilized for the analysis of gut microbiomes, revealing correlations between microorganisms and diseases. It has been observed that dysbiosis are accompanied by decreased species richness and specific reductions in microbial taxa such as Ruminococcaceae and Lactobacillus, while Proteobacteria exhibit an increase (Zheng et al., 2020). The presence of certain microbes and their positive correlation with the occurrence and severity of specific diseases, for example, Escherichia coli and Ruminococcus gnavus in inflammatory bowel disease or Anaerotruncus colihominis, Lachnospira perfectinoschiza, and Ruminococcus callidus in cases of bloating and abdominal pain, suggests the potential use of microbes as bacterial biomarkers for disease diagnosis (Companys et al., 2021; Zhang et al., 2015).
Comprehensive fecal collection is important for multiple purposes, such as individual-specific gut microbiome community research and isolation of beneficial bacteria for the development of probiotics for animals or humans. Typical stool banks intended for fecal microbiota transplantation (FMT) primarily consist of fecal samples from healthy donors (Barnes and Park, 2017), restricting the diversity of intestinal microbial compositions available for research and medical purposes. In contrast, YS Flora® collected fecal samples from both healthy and non-healthy individuals, aiming to gain a more comprehensive understanding of the relationship between gut microbiome communities, diseases, and lifestyles. We collected fecal samples and corresponding information on physical conditions and lifestyles from over 300 donors and named this collection YS Flora®. To demonstrate the application of community analysis of YS Flora® fecal samples, we compared gut microbial communities between vegetarian and omnivorous dietary habits. Additionally, numerous beneficial bacteria with probiotic potential were isolated using selective media. YS Flora® aims to expand knowledge on the gut microbiome and contribute to human health as a multi-purpose collection of human feces and gut microbiome.
Materials and Methods
The fecal collection process was conducted with approval from the Public Institutional Review Board (permission number P01-202306-06-001). Collection papers, tubes, consent forms, and questionnaires were distributed nationwide. For children, consent forms and questionnaires were adjusted to their appropriate level of understanding. Additionally, the questionnaire data was organized in our spreadsheet, which can be provided individually upon request. When categorizing the medication status of donors, we employed the drug classification criteria outlined by the Ministry of Food and Drug Safety (MFDS) of South Korea. Body mass index (BMI) was classified into underweight (< 18.5), normal weight (18.5~24.9), overweight (25.0~29.9), and obesity (30 ≤) categories based on the criteria of the World Health Organization.
For long-term storage, the fecal samples were suspended in glycerol. A sterilized solution of 25% glycerol and 0.9% NaCl (GS solution) was individually autoclaved at 121°C for 15 minutes and then combined. Subsequently, 1 g of feces was suspended in 9 mL of the GS solution, vigorously mixed, and frozen at –80°C.
Six vegetarians and six omnivores were selected from the pool of donors, as dietary habits have been identified as one of the key factors influencing the composition of the intestinal microbial community. Fecal samples were primarily obtained from donors who did not consume probiotics or medications.
DNA extraction from the fecal samples of vegetarians and omnivores was carried out using the PowerSoil® DNA Isolation Kit (Qiagen, CA, USA). The V3-V4 region of the bacterial 16S rDNA was then amplified for subsequent amplicon sequencing, with the extracted DNA serving as the template. PCR was performed using specific primers designed for the DNA region (Forward: 5’-TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG CCT ACG GGN GGC WGC AG-3’; Reverse: 5’-GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA GGA CTA CHV GGG TAT CTA ATC C-3’).
For PCR amplification, a PCR master mix was prepared by combining 12.5 ng of DNA, 1 μM of forward and reverse primers (5 μL each), and 2x KAPA HiFi HotStart Ready Mix (12.5 μL). PCR was conducted under the following conditions: initial denaturation at 95°C for 3 minutes, followed by 25 cycles of denaturation at 95°C for 30 seconds, annealing at 55°C for 30 seconds, elongation at 72°C for 30 seconds, and a final elongation step at 72°C for 5 minutes. The PCR products were purified using AMPure XP beads to remove residual primers and primer dimers.
The purified V3-V4 region amplicon (5 μL) was combined with the Nextera® XT Index Kit to construct the final sequencing library. This involved mounting the sample in a TruSeq Index Plate fixture and adding 5 μL of Index 1 and Index 2 primers from the Nextera XT Index Kit. Subsequently, the second PCR was performed using 2x KAPA HiFi HotStart Ready Mix (25 μL) and PCR Grade Water (10 μL), with the cycle count adjusted to 8 cycles for indexing the DNA library. The index-added V3-V4 region PCR amplicon was purified using AMPure XP beads.
The sample DNA libraries, combined with the attached indexes, were normalized using Qubit (Thermo Fisher Scientific, MA, USA) and Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA). Before cluster creation and sequencing, the DNA libraries underwent denaturation using sodium hydroxide, hybridization buffer, and the PhiX Control Kit. The sequencing was performed using MiSeq FGx (Illumina, CA, USA) equipment.
In the amplicon sequencing results, the read files were processed using MiSeq Reporter software, Illumina’s on-instrument secondary analysis tool. Only sequences meeting the minimum base call quality criteria (Q30) were included for further analysis. The sequence data was then uploaded to the Quantitative Insights into Microbiological Ecology program and demultiplexed based on the tagging short index sequences.
To improve the accuracy of the reads, the Divisive Amplicon Denoising Algorithm 2 was employed for error calibration, and chimera sequences were removed. Subsequently, an amplicon sequence variant (ASV) feature table was generated. The bacterial identification for each ASV was conducted by referencing the 16S rRNA database from the National Center for Biotechnology Information.
Chao1 and Shannon’s indices were utilized to analyze alpha diversity. The beta diversity analysis involved the utilization of the Bray-Curtis dissimilarity metric, which was visualized using an unweighted-pair group method with an average linkage phylogenetic tree and principal coordinates analysis (PCoA) employing the weighted unifrac distance analysis method. A one-way analysis of similarities (ANOSIM) was conducted using the software R’s vegan package with a maximum of 999 permutations to assess the significance of dissimilarity between clusters. This analysis compared the similarity between vegetarians and carnivores in terms of global R and p-values.
Bacteria widely recognized as beneficial, such as Lactobacillus and Bifidobacterium, were isolated from the fecal samples of the donors using selective media. Rogosa agar (Kisan Bio) and Bifidobacterium selective medium (BSM; Sigma) were employed to specifically isolate Lactobacillus spp. and Bifidobacterium spp., respectively. Medium supplements were added to the sterilized media separately.
Bacterial isolation was processed under aerobic or anaerobic conditions. The anaerobic condition in the anaerobic chamber (Vinyl type, Coy Laboratory Products, MI, USA) consisted of mixed gas with an N2:CO2:H2 ratio of 90:5:5. After serial dilution, 100 μL of fecal suspension was spread onto the reduced Rogosa and BSM agar plates and cultured anaerobically at 37°C for 48 hours. Bacillus species were isolated using the tryptic soy agar (TSA) medium under aerobic conditions at 37°C for 24 hours. Colonies with different morphologies were picked with sterile loops, then streaked on de Man, Rogosa, and Sharpe (MRS) agar containing 0.05% L-cysteine hydrochloride for Lactobacillus spp. or Bifidobacterium spp., or on TSA for Bacillus spp.
Polymerase chain reaction (PCR) was conducted to amplify 16S rRNA sequences and identify the individual isolated colonies. The DNA region with a length of 1,500 bp was amplified using the universal primer set 27F 5’-AGA GTT TGA TCC TGG CTC AG-3’ and 1492R 5’-GGT TAC CTT GTT ACG ACT T-3’. A single colony was picked and suspended in 100 μL of sterile water for colony PCR. A 30 μL reaction mixture consisting of 2X TOP simple Dye MIX-Tenuto PCR Prefix (20 μL), 10 pM forward and reverse primers (1 μL each), colony suspension (3 μL), and distilled water (5 μL) was prepared. PCR for 16S rRNA amplification composed of an initial denaturation step of 3 minutes at 94°C, followed by 30 cycles of 30 seconds at 94°C, 30 seconds at 58°C, and 2 minutes at 72°C. A final extension step of 10 minutes at 72°C was performed. The PCR product was purified using the Mini BEST DNA Fragment Purification Kit (TaKaRa Bio, Japan) and subsequently sequenced at Bionics (Korea).
Results
To ensure long-term storage, the feces were mixed with glycerol and frozen in a deep freezer. Based on age distribution, individuals in their 60s constituted the largest group, followed by those in their 50s (Fig. 1A). Regarding BMI, the majority (56%) fell into the normal weight (18.5~24.9) category. In comparison, only 8% were underweight (< 18.5), and 4% were classified as obese (30 ≤) category (Fig. 1B). In terms of caffeine and alcohol consumption, the majority of donors (60%) reported consuming less than one drink per week (Figs. 1C and 1D). Additionally, the number of individuals adhering to a vegetarian diet was 1.5 to 2 times higher than that of omnivores (Fig. 1E). An overwhelming majority (89%) reported defecating up to two times a day (Figs. 1F).
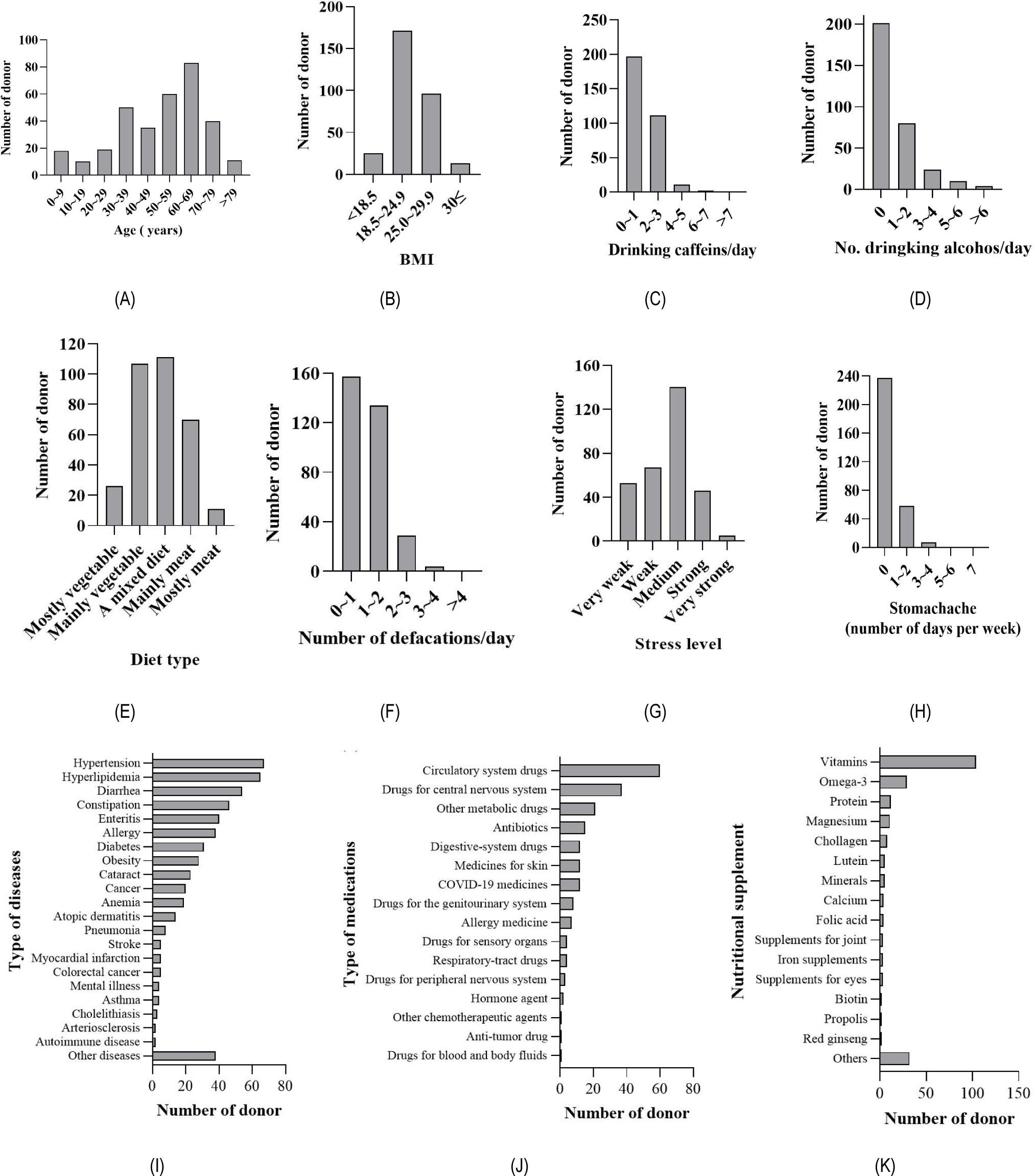
Stress levels among donors were primarily reported as moderate or weaker (83%), and 78% experienced zero episodes of daily abdominal pain (Figs. 1G and 1H). Among the listed symptoms in Figure 1I, approximately 64% of the donors reported experiencing at least one of them, with hypertension being the most prevalent condition based on the questionnaire responses. Among the various types of drugs classified according to the drug classification system of the MFDS of Korea, circulatory system drugs were the most consumed (Fig. 1J). Furthermore, within the healthy and symptomatic donor groups, 56% reported taking antibiotic-containing drugs three months before fecal donation, and multivitamins were the most frequently consumed nutritional supplement (Fig. 1K).
Research on the human gut microbiome community can be facilitated by utilizing fecal samples from YS Flora®. As an example, we conducted a metagenomic analysis and comparison of the gut microbiomes of donors following vegetarian and omnivorous diets. The sequencing depth for all samples exceeded 50,000 reads, with an average of 178,406 reads. After undergoing quality control, a total of 2,140,872 reads were obtained. The vegetarian group yielded a confirmed total of 1,092,614 reads, while the omnivore group had 1,048,258 reads. On average, each sample in the vegetarian group contained 182,102 reads, while the omnivore group had an average of 174,710 reads per sample. With an average Good’s coverage of 99.97%, the sequencing depth achieved by our read counts ensures a comprehensive capture of microbial diversity, indicating a high probability that we have sequenced the vast majority of species in our samples.
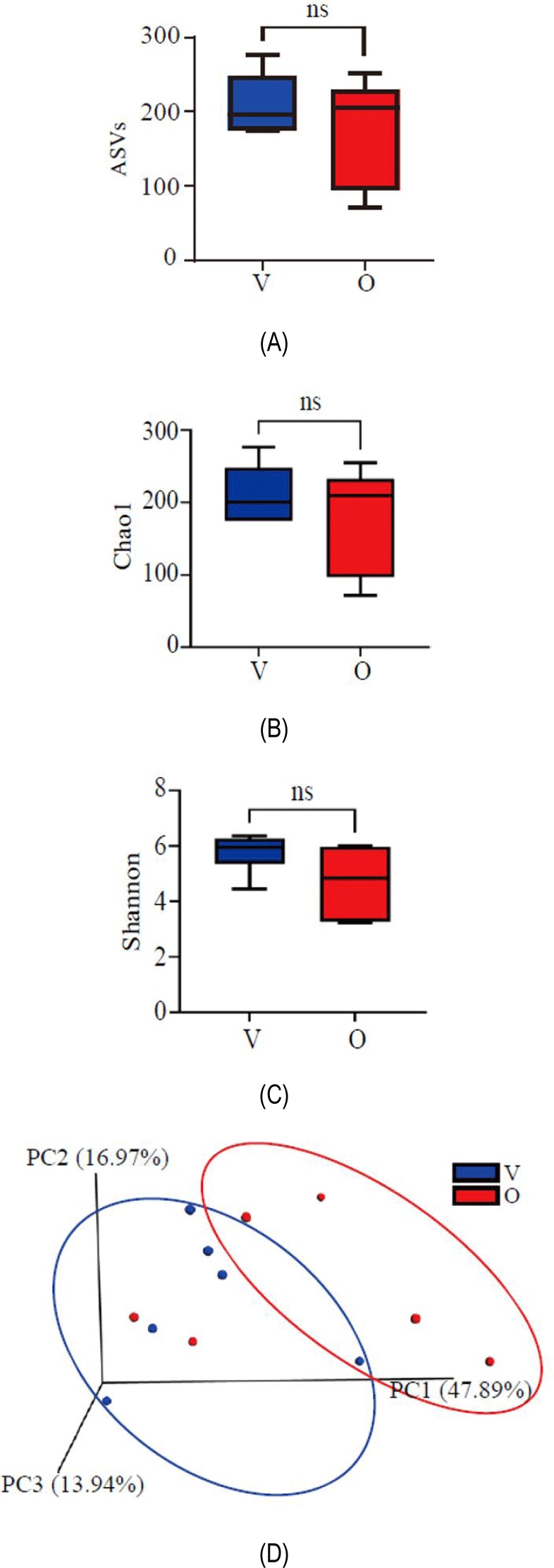
The alpha diversity analysis between the two dietary groups was represented by ASVs, Chao1 and Shannon indices (Figs. 2A-C). Statistical analysis revealed no significant difference in the average number of ASVs between the vegetarian and the omnivorous group. The Chao1 index, which reflects the number of unique ASVs in the community, showed a higher value in the vegetarian groups, although the difference was not statistically significant. The Shannon index, which measures microbial community diversity, showed a slight increase in the vegetarian group compared to the omnivore group, indicating greater diversity in individuals following a vegetable-oriented diet, but the difference did not reach statistical significance. To evaluate the similarity of bacteria composition between samples, we performed beta diversity analysis using the weighted unifrac method and visualized the results using PCoA (Fig. 2D). Microbiome compositions were found to be better maintained among vegetarians when compared to those of omnivores. The comparison using the Bray-Curtis similarity index and ANOSIM indicated that the differences in gut microbiota composition between the two groups were not statistically significant (global R = 0.02037, p = 0.3487).
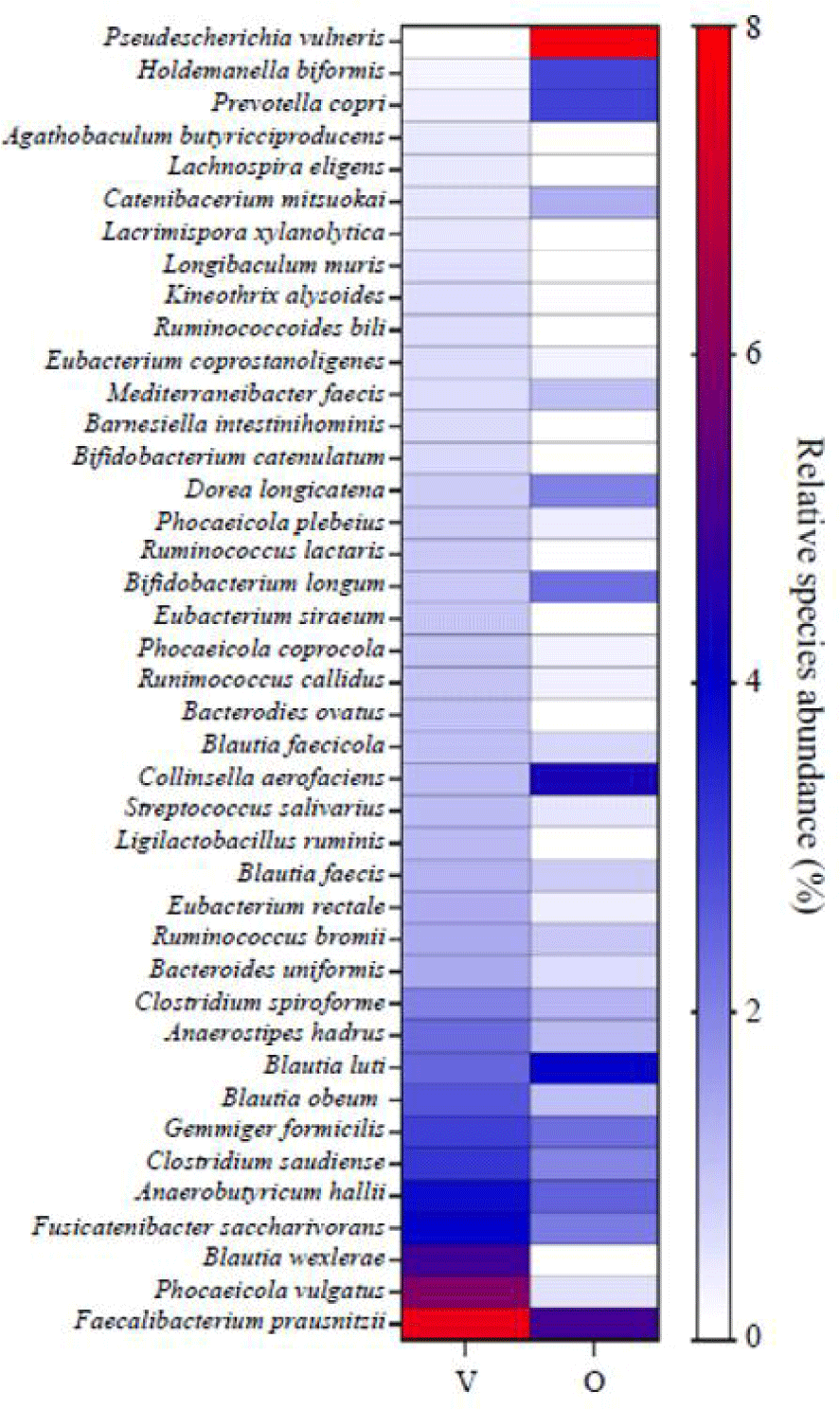
The composition of abundant species was compared between vegetarians and omnivores, resulting in a total of 372 species being analyzed. The microbiome compositional structures differ significantly between the two groups. The relative abundance at the species level was ranked in ascending order for the vegetarian group (Fig. 3). The top three species in the vegetarian group were Faecalibacterium prausnitzii, Phocaeicola vulgatus, and Blautia wexlerae. F. prausnitzii constituted an average of 7.5%, while the top three strains accounted for 18.4%. In the omnivore group, Pseudescherichia vulneris, Collinsella aerofaciens, and Blautia luti were found to be enriched. Ps. vulneris accounted for 7.9%, and the combined abundance of the top three strains was 13.7%.
To isolate specific strains of Lactobacillus spp. and Bifidobacterium spp., including those recognized for overall safety by the MFDS of Korea and other potentially beneficial strains for probiotic or pharmabiotic development, we utilized a fecal-glycerol suspension on two selective media and one basic medium. A total of 878 strains were isolated from the YS Flora®. Among them, 400 isolates belong to the species designated by the MFDS, meaning their overall safety is recognized and guaranteed by the Korean MFDS, allowing for their commercialization without additional safety evaluations (Table 1 and Supplementary Table S1). In addition to the MFDS-designated strains, a total of 405 species from which more than 10 isolates were obtained and which have been reported in research to have health-promoting or disease-curing functionalities are listed in Table 2. Bacillus spp. was predominantly isolated from the basic medium TSA. Lactic acid bacteria such as Lactobacillus spp., Weissella cibaria, Pediococcus spp., and Leuconostoc mesenteroides were primarily isolated from the Rogosa medium.
Discussion
The main objective of this study was to advance the utilization of fecal collection YS Flora® for multiple applications, encompassing the isolation of microbes for the development of probiotics or live biotherapeutic products, as well as investigations into the composition of the gut microbiome. The donor information includes age, BMI, alcohol and caffeine consumption, dietary habits, stool frequency, stress level, abdominal pain, diseases, medication, and supplement (Figs. 1A-K), documented in our spreadsheet. YS Flora® provides a collection of fecal samples from a wide range of age groups, enabling analysis of various studies such as the age-related core microbiome composition and variations in short-chain fatty acid-producing bacteria (Odamaki et al., 2016). Moreover, YS Flora® is well-suited for comparing gut microbial communities between individuals with a healthy condition and those with 22 different diseases and disorders (Fig. 1I). Typical stool banks operating for FMT purposes select only a small percentage (less than 5%) of fecal samples from healthy donors after rigorous testing (He et al., 2021). In contrast, YS Flora®, not intended for FMT, has also secured fecal samples from non-healthy individuals. This approach allows insights into the associations between gut microbial composition and health conditions and lifestyles, such as stress levels, antibiotics usage, stomachaches, and diseases (De Vos et al., 2022).
The community analysis results of fecal samples from vegetarians and omnivores demonstrate that 16S rRNA metagenomic amplicon sequencing of YS Flora® fecal samples can be represented in terms of alpha or beta diversity and predominant species (Figs. 2A-D). Although this example did not show significant differences in alpha and beta diversity between vegetarian and omnivore feces, these are not always similar and can vary depending on nutrient intake, including carbohydrates, proteins, and fats, despite the contrasting diets or other factors such as age, sex, and BMI (Losasso et al., 2018; Matijašić et al., 2014; Ruengsomwong et al., 2016). In the analysis of dominant species within each dietary group, F. prausnitzii, P. vulgatus, and B. wexlerae were identified as the predominant species in the fecal samples of vegetarians, which aligns with previous studies where these species were commonly found in the feces of vegetarians and considered indicative of a vegetarian diet (Dridi et al., 2023; Ferrocino et al., 2015; Kjølbæk et al., 2020; Nakajima et al., 2020; Patnode et al., 2019; Takei et al., 2022; Xia et al., 2022; Zafar and Saier, 2021). On the other hand, among the abundant species in the fecal samples of the omnivore group, P. vulneris, C. aerofaciens, and B. luti were prominently observed. C. aerofaciens has been reported to be more prevalent in the non-vegetarian group and in individuals with low dietary fiber intake (Gomez-Arango et al., 2018; Ruengsomwong et al., 2016). However, further research is required to elucidate the reasons behind the higher abundance of P. vulneris, a microorganism widely distributed in different parts of the human body (Brenner et al., 1982), and B. luti, known for its ability to ferment carbohydrates (Kjølbæk et al., 2020), in the fecal samples of individuals following a meat-based diet.
Species identified as designated strains of MFDS and others, with more than 10 isolates from YS Flora® feces, with potential health benefits or disease healing properties based on literature research. The strains listed in Table 1 are considered relatively safe and can potentially be developed into probiotics and therapeutic agents based on their confirmed functionality without additional safety verification (Fijan, 2014). Over 400 strains with various potential functionalities have been secured (Table 2), among which Bacillus species have been reported to exhibit antimicrobial activity, antiviral effects, and toxin neutralization (Byun et al., 2023; Lee et al., 2019; Xie et al., 2023; Zhang et al., 2021). Lactobacillus sakei and Lactobacillus brevis have been studied for increasing beneficial microbial richness and abundance, metabolic regulation and anti-diabetic properties, anti-inflammatory effects, and immune regulation (Chen et al., 2023; Kwon et al., 2018; Riccia et al., 2007; Zou et al., 2023). Pediococcus strains have been reported for metabolic regulation, proliferation of beneficial microbes, and suppression of harmful pathogens (Al-Emran et al., 2022; Silva et al., 2017; Ueda et al., 2018). W. cibaria and Leu. mesenteroides have been found to improve oral health by reducing halitosis and inhibiting Streptococcus mutans, and to alleviate skin conditions such as psoriasis and atopic dermatitis (Lee et al., 2021; Lim et al., 2017; Luan et al., 2022; Ogawa et al., 2021). Bifidobacterium pseudocatenulatum has been studied for enhancing intestinal barrier function, reducing intestinal permeability, and consequently decreasing bacteria-induced inflammatory responses, such as in rheumatoid arthritis and cirrhosis (Moratalla et al., 2016; Zhao et al., 2023).
YS Flora® is a comprehensive collection of human feces from individuals with diverse lifestyles and physical conditions. This collection provides a representative sample of an individual’s intestinal microbial pool for multiple purposes, such as community analyses, and secures isolates of beneficial microbes. Further studies will evaluate the safety and efficacy of beneficial microbes derived from YS Flora® for specific indications. Furthermore, conducting community analyses with a larger sample size of fecal samples will contribute to expanding our knowledge of the relationship between gut microbiota, lifestyles, and diseases.
