Research Article
In Vitro Assessment of Probiotic Properties for Lactic Acid Bacteria Isolated from Korean Traditional Fermented Food, Kimchi
Eui-Sang Cho1
,
Jeonghye Chun1
,
Jeongin Park2
,
Mibang Kim1
,
Chi Young Hwang1
,
Deok Jun Yoon1
,
Inonge Noni Siziya3
,
Dong-Ho Seo3,*
,
Myung-Ji Seo1,2,*
Author Information & Copyright ▼
1Department of Bioengineering and Nano-Bioengineering, Graduate School of Incheon National University, Incheon 22012, Korea
2Division of Bioengineering, Incheon National University, Incheon 22012, Korea
3Department of Food Science and Technology, College of Agriculture and Life Sciences, Jeonbuk National University, Jeonju 54896, Korea
*Corresponding author : Dong-Ho Seo, Department of Food Science and Technology, College of Agriculture and Life Sciences, Jeonbuk National University, Jeonju 54896, Korea. Tel: +82-63-270-2571, E-mail:
dhseo@jbnu.ac.kr
*Corresponding author : Myung-Ji Seo, Division of Bioengineering, Incheon National University, Incheon 22012, Korea. Tel: +82-32-835-8267, E-mail:
mjseo@inu.ac.kr
Copyright © Korean Society for Lactic Acid Bacteria and Probiotics. This is an Open-Access article distributed under the terms of the
Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0/) which permits
unrestricted non-commercial use, distribution, and reproduction in any
medium, provided the original work is properly cited.
Received: May 31, 2020; Revised: Jun 12, 2020; Accepted: Jun 14, 2020
Published Online: Jun 30, 2020
Abstract
In this study, thirty-eight lactic acid bacteria (LAB) strains were isolated from Kimchi (Korean traditional fermented food) and screened for probiotic characterization in a series of in vitro tests, including resistance to low pH, tolerance to bile salts, hydrophobicity, antibiotic resistance and antioxidant activity. Firstly, twenty-two strains displayed survival in acid and bile passage conditions. Secondly, the strains KCCP 11349 and KCCP 11356 were observed to have high cell surface hydrophobicity with 63.9 and 98.8 %, respectively. Functionality of these potential probiotic isolates was supported by their antioxidant activity. Altogether, the strain KCCP 11349 showed characteristics similar to or even better than reference strain Lactobacillus rhamnosus GG. Finally, 16S rRNA gene sequencing was conducted to identify potential probiotic strains and KCCP 11349 was identified as Lactobacillus plantarum subsp. plantarum.
Keywords: Kimchi; lactic acid bacteria; probiotic
Introduction
Nowadays, there is considerable interest in functional foods with regards to their beneficial effects on human health. Accordingly, the preference for foods containing probiotics is constantly increasing, mainly due to the numerous publications made that have linked them to the promotion of positive effects on health (Aureli et al., 2011).
Kimchi is a Korean traditional fermented food classified as one of the healthiest foods as it is rich in vitamins, and has a variety of beneficial effects such as aiding in digestion, and cancer prevention. Several researchers have previously isolated and studied various probiotics from kimchi, particularly Lactic acid bacteria (LAB) (Choi et al., 2002; Hwang et al., 2000; Kim et al., 2002). With regards to LAB and other ingredients used in Kimchi, the nutritional, functional and organoleptic profiles change as fermentation progresses and when consumed, kimchi helps to restore the probiotic microbiota in the gut.
Probiotics are officially defined as: ‘‘live microorganisms which, when administered in adequate amounts, exert a beneficial effect on the health of the consumer” (FAO/WHO, 2002). Probiotics can modulate host immune responses to promote a healthy gastrointestinal tract. LAB are the most prominent microorganisms applied as probiotics (Rivera-Espinoza and Gallardo-Navarro, 2010) and they have been studied as sources of antibacterial, anticancer (Shin et al., 1998), anti-diabetic (Yadav et al., 2007), anti-obesity (Arora et al., 2013; Kang et al., 2013) and anti-oxidant (Das and Goyal, 2015; Spyropoulos et al., 2011) components. The antioxidant activity of some LAB used as food components and probiotics may have a substantial impact on human health (Lin et al., 2000, Oxman et al., 2000).
To assess such possibilities, the aim of this study was to isolate potential probiotic strains from kimchi which show probiotic characterization and evaluate them in a series of in vitro tests, including low pH resistance, tolerance to bile salts, hydrophobicity, resistance to antibiotics and antioxidant activity.
Materials and Methods
Isolation and identification of bacteria from kimchi
Homemade kimchi was collected and serial dilutions were made in saline. The saline mixtures were spread on de Man, Rogosa and Sharpe (MRS) agar followed by overnight incubation at 30°C. To obtain a pure colony, a single representative colony was transferred to a new MRS agar plate at least three times. The pure isolates were preserved at –80°C in 25% (w/v) glycerol suspension for long-term storage.
Acid and bile salt tolerance
To determine the tolerance level of isolated LAB in low pH conditions and bile salt conditions we used the method described by Pieniz et al. (2004) with slight modifications. An overnight culture of potential probiotics was centrifuged at 12,000 ×g for 5 min at 4°C. The cell pellet washed with 0.85% NaCl (w/v) solution and suspended in 0.85% NaCl (w/v) solution adjusted to pH 2.0, prior to being incubated at 37°C for 2 h. After incubation viable cell count was determined on MRS agar plate. For bile salt tolerance, a similar protocol was followed. Cell pellet was dissolved in 0.85% NaCl (w/v) solution supplemented with 1.5% and 2.0% bile salt (BD) and further incubated at 37°C for 4 h. After incubation the cell viable count was determined on MRS agar plate. The strains with acid and bile salt tolerance were primarily selected and the following experiments were conducted on the selected strains.
Bacterial adhesion to hydrocarbons (BATH)
BATH test was performed according to an established protocol with simple modifications (Collado et al., 2007). Bacterial cells were washed with 0.85% NaCl (w/v) solution and re-suspended in the same solution. Initial OD600 adjusted to 0.2 to 0.3. One milliliter of toluene was added to bacterial cells and vortexed well for about 5 min, prior to being incubated for 1 h at room temperature. When two distinct phases were observed, the aqueous phase was removed and the reading was noted. BATH% was calculated as follows:
where A and A0 represent absorbance readings after and before mixing with toluene.
Antibiotic susceptibility test
The antibiotic susceptibilities of strains isolated from kimchi were tested using paper disc diffusion methodology (Bauer et al., 1996). Overnight culture of isolates was spread onto the MRS agar plate and discs containing specific antibiotic concentrations were placed onto them. The plates were incubated overnight at 37°C for 24 h and the inhibition zones were identified. Antibiotic concentrations used in this experiment were as follows: Ampicillin (100 μg/mL), ciprofloxacin (10 μg/mL), erythromycin (25 μg/ mL), gentamicin (30 μg/mL), lincomycin (15 μg/mL), novobiocin (10 μg/mL), streptomycin (50 μg/mL) and tetracyclin (30 μg/mL).
Antioxidant properties of LAB isolates in MRS broth
The isolates were inoculated into 20 mL of MRS broth (n=3) and incubated overnight at 37°C. After centrifugation (15,000 ×g for 10 min at 4°C), the scavenging activity of the isolates was determined using the supernatant. 2,2-diphenyl-1-picrylhydrazyl (DPPH) was added to the supernatant and the antioxidant activity was quantified using a colorimetric microplate assay described by Kim et al. (2019). Prepared DPPH solution (0.2 mM in ethanol; Sigma Aldrich, USA) was mixed with bacterial supernatant. The reaction mixture was incubated in the dark at room temperature for 30 min. The DPPH radical scavenging activity was then detected by measuring the absorbance at 517 nm using a microplate reader. The scavenging ability was calculated as follows:
Identification of selected LAB
Identification of isolates was performed at the 16S rRNA gene sequencing facility of Macrogen Co., Ltd. (Seoul, Korea). The obtained DNA sequence was analyzed by using BLAST program provided by GenBank (https://www.ncbi.nlm.nih.gov/) and EzBioCloud server (http://www.ezbiocloud.net/) (Yoon et al., 2017) for phylogenetic analysis. Phylogenetic analysis was conducted by MEGA 7 (Kumar et al., 2016) after a gap deletion and multiple alignments of data via the CLUSTAL W program (Thompson et al., 1994). The evolutionary distances were calculated using the Kimura two-parameter model (Kimura, 1980), and neighbor-joining (NJ) algorithm was used to construct the phylogenetic trees.
Results and Discussion
Isolation and screening of probiotic strains with acid and bile salt resistant
Thirty-eight isolates of LAB isolated from kimchi were tested for their abilities to resist high acidity at the low pH of 2.0 and respective bile salt concentrations of 1% and 2% (w/v). All isolates were observed to have survival rates above 75% with tolerance to 1% and 2% bile salt (w/v), and of the isolates that were screened for tolerance to pH 2.0, twenty-two had an adequate survival rate of between 20 and 60% (Table 1). The bile and pH tolerance of microbiota within the gastric juice are considered among the main factors affecting the survival of probiotic bacteria upon passage through the stomach to the intestine. Interestingly, all of the strains showed high survival rate in bile salts at 1% and 2%, particularly the strain KCCP 11349, which also presented adequate viability in pH 2 and was comparable to the reference strain. Collectively, twenty-two strains of LAB recorded probiotic potential and were selected for further study.
Table 1.
Assessment of viability of the isolated strains after exposure to low pH and bile salts. In all tests, the probiotic L. rhamnosus GG KCTC 5033 served as a reference strain
| Number |
Strains |
Initial counts |
After 2 h (pH 2.0) |
After 4 h (Bile salts 1%, w/v) |
After 4 h (Bile salts 2%, w/v) |
| Log (cfu/mL) |
Survival (%) |
Log (cfu/mL) |
Survival (%) |
Log (cfu/mL) |
Survival (%) |
| 1 |
LGG |
7.5 |
4.5 |
60.0 |
6.8 |
90.8 |
6.7 |
89.1 |
| 2 |
JK226 |
9.4 |
5.6 |
59.5 |
9.2 |
97.7 |
9.0 |
96.2 |
| 3 |
JK756 |
9.0 |
5.5 |
60.9 |
7.6 |
85.1 |
7.7 |
85.5 |
| 4 |
KCCP11031 |
9.0 |
3.1 |
34.6 |
8.3 |
92.2 |
8.3 |
92.0 |
| 5 |
KCCP11156 |
10.6 |
4.6 |
43.4 |
9.2 |
86.3 |
9.4 |
88.8 |
| 6 |
KCCP11178 |
9.3 |
2.0 |
20.9 |
9.1 |
97.0 |
9.0 |
96.3 |
| 7 |
KCCP11179 |
9.4 |
0.0 |
0.0 |
9.0 |
95.6 |
9.1 |
97.2 |
| 8 |
KCCP11180 |
9.5 |
0.0 |
0.0 |
9.2 |
97.4 |
9.3 |
98.6 |
| 9 |
KCCP11181 |
9.4 |
0.0 |
0.0 |
9.2 |
98.7 |
9.2 |
98.7 |
| 10 |
KCCP11183 |
9.3 |
2.5 |
26.6 |
8.8 |
94.6 |
9.0 |
96.1 |
| 11 |
KCCP11184 |
9.4 |
2.2 |
23.5 |
8.9 |
94.8 |
9.0 |
95.5 |
| 12 |
KCCP11186 |
9.3 |
2.4 |
26.2 |
8.8 |
94.8 |
8.7 |
93.1 |
| 13 |
KCCP11209 |
9.6 |
0.0 |
0.0 |
8.9 |
93.3 |
7.9 |
82.3 |
| 14 |
KCCP11213 |
9.3 |
1.7 |
17.9 |
8.9 |
96.0 |
9.0 |
96.8 |
| 15 |
KCCP11218 |
9.3 |
1.1 |
12.1 |
9.0 |
96.9 |
9.1 |
98.2 |
| 16 |
KCCP11219 |
9.3 |
0.0 |
0.0 |
9.1 |
98.4 |
9.1 |
98.5 |
| 17 |
KCCP11222 |
9.4 |
1.1 |
12.0 |
9.1 |
97.4 |
8.8 |
94.2 |
| 18 |
KCCP11223 |
8.4 |
2.3 |
27.9 |
7.7 |
91.8 |
7.8 |
93.1 |
| 19 |
KCCP11298 |
9.1 |
0.0 |
0.0 |
8.7 |
94.8 |
8.6 |
94.4 |
| 20 |
KCCP11303 |
9.3 |
3.0 |
32.1 |
8.5 |
91.4 |
8.6 |
91.7 |
| 21 |
KCCP11308 |
9.2 |
0.5 |
5.7 |
8.9 |
96.7 |
8.9 |
96.8 |
| 22 |
KCCP11309 |
9.2 |
5.1 |
55.6 |
8.8 |
95.3 |
8.9 |
96.5 |
| 23 |
KCCP11316 |
9.1 |
0.0 |
0.0 |
7.9 |
87.0 |
7.9 |
86.9 |
| 24 |
KCCP11317 |
9.4 |
2.8 |
29.6 |
8.7 |
92.9 |
8.6 |
92.4 |
| 25 |
KCCP11318 |
9.1 |
3.1 |
34.5 |
8.1 |
89.9 |
8.2 |
90.5 |
| 26 |
KCCP11319 |
9.3 |
4.6 |
50.0 |
8.9 |
96.4 |
9.0 |
96.6 |
| 27 |
KCCP11324 |
8.2 |
1.8 |
22.1 |
7.9 |
95.4 |
8.0 |
97.3 |
| 28 |
KCCP11349 |
9.3 |
5.8 |
62.4 |
8.9 |
95.6 |
8.7 |
93.3 |
| 29 |
KCCP11354 |
9.0 |
3.9 |
43.4 |
8.8 |
97.7 |
8.8 |
97.9 |
| 30 |
KCCP11355 |
9.1 |
0.0 |
0.0 |
8.9 |
98.2 |
8.7 |
95.9 |
| 31 |
KCCP11356 |
9.0 |
3.9 |
43.4 |
8.8 |
97.7 |
8.8 |
97.9 |
| 32 |
KCCP11370 |
8.7 |
1.0 |
11.5 |
8.2 |
94.0 |
7.8 |
89.4 |
| 33 |
KCCP11376 |
10.2 |
0.5 |
5.1 |
8.0 |
78.6 |
7.8 |
75.9 |
| 34 |
KCCP11446 |
9.3 |
4.3 |
46.0 |
8.8 |
94.5 |
9.0 |
96.2 |
| 35 |
66 |
9.0 |
3.6 |
40.3 |
8.7 |
97.1 |
8.6 |
96.2 |
| 36 |
70 |
9.3 |
4.8 |
51.8 |
8.7 |
93.7 |
8.7 |
93.9 |
| 37 |
75 |
8.5 |
0.0 |
0.0 |
7.8 |
92.3 |
7.7 |
91.3 |
| 38 |
78 |
8.2 |
2.2 |
27.1 |
6.8 |
82.7 |
6.8 |
82.4 |
| 39 |
115 |
9.2 |
0.0 |
0.0 |
7.7 |
83.6 |
7.6 |
82.9 |
Download Excel Table
Bacterial adhesion to hydrocarbon (Hydrophobicity)
The selected strains on the basis of their survival under in vitro GIT conditions, were evaluated for their hydrophobicity toward toluene that may reflect the colonization potential of the organism in the intestinal lumen. The degree of hydrophobicity was high for KCCP 11356 with the highest hydrophobicity value of 98.8% (Table 2). This value was significantly higher than Lactobacillus rhamnosus GG (82.8%). The hydrophobicity of the outer surface of microorganisms has been implicated in the attachment of bacteria to host tissue. This property may provide an important competitive advantage in maintaining bacteria in the human gastrointestinal tract (Singh et al., 2012). Several tested strains exhibited weak hydrophobicity values for toluene, while hydrophobicity values of KCCP 11349 and KCCP 11356 presented strong hydrophobicity (63.9± 0.6 and 98.8±0.3%, respectively). The variation in hydrophobicity has been reported in other probiotic strains and has been elucidated by the fact that attachment depends upon the origin of the strains as well as their surface properties (Abushelaibi et al., 2017).
Table 2.
Percent hydrophobicity of selected strains to toluene
| Number |
Strains |
Hydrophobicity (%) |
| 1 |
LGG |
82.8±0.3 |
| 2 |
JK226 |
6.5±3.0 |
| 3 |
JK756 |
0.1±0.1 |
| 4 |
KCCP11031 |
3.1±1.3 |
| 5 |
KCCP11178 |
1.6±1.0 |
| 6 |
KCCP11183 |
4.7±1.4 |
| 7 |
KCCP11184 |
1.3±0.6 |
| 8 |
KCCP11186 |
0.4±0.2 |
| 9 |
KCCP11213 |
0.7±0.7 |
| 10 |
KCCP11223 |
0.4±0.2 |
| 11 |
KCCP11226 |
96.8±0.8 |
| 12 |
KCCP11303 |
2.3±1.9 |
| 13 |
KCCP11309 |
1.8±1.4 |
| 14 |
KCCP11317 |
2.6±1.4 |
| 15 |
KCCP11318 |
0.2±0.1 |
| 16 |
KCCP11319 |
3.5±2.2 |
| 17 |
KCCP11324 |
2.5±1.2 |
| 18 |
KCCP11349 |
63.9±0.6 |
| 19 |
KCCP11354 |
7.0±1.0 |
| 20 |
KCCP11356 |
98.8±0.3 |
| 21 |
KCCP11446 |
6.1±0.2 |
| 22 |
66 |
18.7±0.3 |
| 23 |
70 |
2.4±1.0 |
| 24 |
78 |
17.1±3.3 |
Download Excel Table
Antibiotic susceptibility test
The selected strains were resistant to erythromycin but only four strains were resistant to novobiocin (Table 3).
Table 3.
Antibiotic susceptibility profiles of potential probiotic strain
| Number |
Strains |
Antibiotics resistant |
| Amp |
Ery |
Gen |
Cip |
Lin |
Nov |
Tet |
Strep |
| 1 |
LGG |
Sa |
Rb |
S |
S |
S |
S |
S |
S |
| 2 |
JK226 |
S |
R |
R |
R |
R |
S |
R |
R |
| 3 |
JK756 |
S |
R |
R |
R |
R |
S |
R |
S |
| 4 |
KCCP11031 |
S |
R |
R |
R |
S |
S |
R |
S |
| 5 |
KCCP11178 |
R |
R |
R |
R |
S |
R |
S |
R |
| 6 |
KCCP11183 |
R |
R |
R |
R |
S |
S |
S |
R |
| 7 |
KCCP11184 |
R |
R |
R |
R |
S |
S |
S |
R |
| 8 |
KCCP11186 |
S |
R |
R |
R |
S |
R |
R |
R |
| 9 |
KCCP11213 |
R |
R |
R |
R |
S |
S |
S |
S |
| 10 |
KCCP11223 |
S |
R |
R |
R |
S |
R |
R |
R |
| 11 |
KCCP11303 |
R |
R |
R |
R |
S |
S |
S |
S |
| 12 |
KCCP11309 |
R |
R |
S |
R |
S |
S |
S |
S |
| 13 |
KCCP11317 |
R |
R |
R |
R |
S |
S |
S |
R |
| 14 |
KCCP11318 |
S |
R |
R |
R |
S |
R |
S |
R |
| 15 |
KCCP11319 |
R |
R |
R |
R |
S |
S |
S |
S |
| 16 |
KCCP11324 |
S |
R |
R |
R |
S |
R |
R |
R |
| 17 |
KCCP11349 |
S |
R |
R |
R |
R |
S |
R |
R |
| 18 |
KCCP11354 |
S |
R |
R |
R |
R |
S |
S |
S |
| 19 |
KCCP11356 |
S |
R |
R |
R |
S |
S |
R |
S |
| 20 |
KCCP11446 |
S |
R |
R |
R |
S |
S |
S |
S |
| 21 |
66 |
S |
R |
S |
S |
S |
S |
R |
S |
| 22 |
70 |
S |
R |
S |
S |
S |
S |
S |
S |
| 23 |
78 |
S |
R |
S |
S |
S |
S |
S |
S |
Download Excel Table
While the resistances to ampicillin and gentamicin was similar to that of the LAB isolates reported previously (Argyri et al., 2013), the other antibiotics showed different patterns for each strain.
Antioxidant properties of LAB isolates in MRS broth
The DPPH-scavenging activity of the strains is shown in Table 4. With the exception of JK226, the potential probiotic strains had a higher antioxidant potential when compared to the reference strain (L. rhamnosus GG). The maximum DPPH radical inhibition activity was observed the strain KCCP 11303 (86.2±1.4). From the above results we could conclude that of the antioxidant potential of probiotic strains depends on the type of strain. These results imply that the metabolic products of LAB might be contributing to the higher antioxidant potential.
Table 4.
Antioxidant activity of potential probiotic strain
| Number |
Strains |
DPPH radical scavenging activity |
| 1 |
LGG |
53.7±2.6 |
| 2 |
JK226 |
51.6±4.1 |
| 3 |
JK756 |
83.4±0.9 |
| 4 |
KCCP11031 |
78.5±7.4 |
| 5 |
KCCP11178 |
65.9±7.1 |
| 6 |
KCCP11183 |
69.6±12.5 |
| 7 |
KCCP11184 |
75.9±4.4 |
| 8 |
KCCP11186 |
76.3±2.9 |
| 9 |
KCCP11213 |
75.6±6.4 |
| 10 |
KCCP11223 |
80.2±2.8 |
| 11 |
KCCP11303 |
86.2±1.4 |
| 12 |
KCCP11309 |
82.3±1.9 |
| 13 |
KCCP11317 |
75.5±3.4 |
| 14 |
KCCP11318 |
83.8±6.6 |
| 15 |
KCCP11319 |
83.9±4.2 |
| 16 |
KCCP11324 |
82.1±3.1 |
| 17 |
KCCP11349 |
57.4±4.7 |
| 18 |
KCCP11354 |
63.1±4.0 |
| 19 |
KCCP11356 |
57.4±5.4 |
| 20 |
KCCP11446 |
58.4±2.0 |
| 21 |
66 |
61.5±6.7 |
| 22 |
70 |
80.3±5.6 |
| 23 |
78 |
77.4±0.9 |
Download Excel Table
Identification of selected LAB by 16S RNA gene sequencing
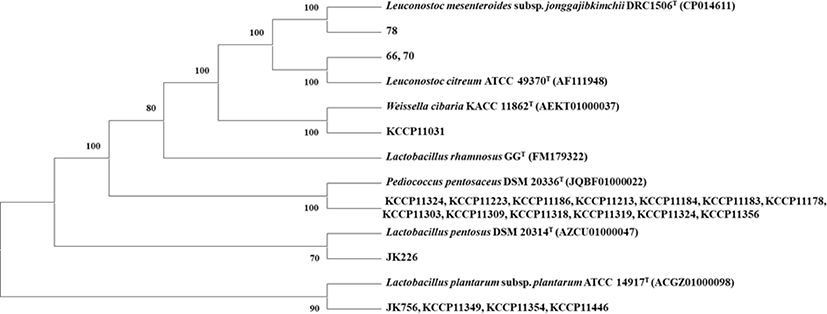
Twenty-two potential probiotics were identified by 16S rRNA gene sequence. The isolates underwent molecular phylogeny analysis and phylogenic tree construction to identify the LAB to a species level based on the 16S rDNA sequences from evolutionary distances by neighbor-joining method. The phylogenetic tree of the 22 isolates with reference strain is shown in Fig. 1. Almost all the isolates belonged to the genera Pediococcus and Lactobacillus, while some belonged to Leuconostoc and Weissella (Fig. 1).
Fig. 1.
Neighbor-joining phylogenetic tree, based on the 16S rRNA gene sequences, showing the position and relationship of strains isolated from kimchi with probiotic potential. Numbers at nodes indicate bootstrap values (>70%) calculated based on the NJ algorithms for the branch point based on 1000 replications. Bar, 0.01 substitutions per nucleotide position.
Download Original Figure
Conclusion
Thirty eight of LAB strains isolated from kimchi were evaluated for their potential probiotic properties. Twenty-two strains were selected with respect to their bile salts and low pH tolerance, and analyzed for their antibiotic susceptibility, and antioxidant activity. Our results indicated that one of the strains, identified by phylogenetic analysis as Lactobacillus plantarum subsp. plantarum KCCP 11349, possessed desirable probiotic properties, which were close to, and in some cases better than, the commercial probiotic L. rhamnosus GG. However, according to the Guidelines for the Evaluation of Probiotics in Food by FAO/WHO (FAO/WHO, 2002), it is necessary to conduct additional in vitro and in vivo experiments to further establish its probiotic character. Also, further studies are required to explore the health benefit of this strain in fermented foods made with this isolate.
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MIST) (No. NRF-2019R1A2C1006038). This research was also supported by the OTTOGI HAM TAIHO FOUNDATION.
References
Abushelaibi A, Al-Mahadin S, El-Tarabily K, Shah NP, and Ayyash M (2017) Characterization of potential probiotic lactic acid bacteria isolated from camel milk.
LWT-Food Sci. Technol.79, 316-325.


Argyri AA, Zoumpopoulou G, Karatzas KAG, Tsakalidou E, Nychas GJE, Panagou EZ, Tassou CC (2013) Selection of potential probiotic lactic acid bacteria from fermented olives by
in vitro tests.
Food Microbiol.
33, 282-291.



Arora T, Singh S, and Sharma RK (2013) Probiotics: Interaction with gut microbiome and antiobesity potential.
Nutrition29, 591-596.



Aureli P, Capurso L, Castellazzi AM, Clerici M, Giovannini M, Morelli L, Poli A, Pregliasco F, Salvini F, Zuccotti GV (2011) Probiotics and health: An evidence-based review.
Pharm. Res.63, 366-376.



Bauer AW, Kirby MM, Sherris JC, and Truck M (1966) Antibiotic susceptibility testing by a standardized single disk method.
Am. J. Clin. Pathol.45, 493-496.



Choi SM, Jeon YS, Rhee SH, and Park KY (2002) Fermentation characteristics and antimutagenicity of kimchi that prepared with different ratio of seed in red pepper powder.
Cancer Res. Treat7, 51-59.

Deeplina D and Arun G (2015) Antioxidant activity and
γ-aminobutyric acid (GABA) producing ability of probiotic
Lactobacillus plantarum DM5 isolated from Marcha of Sikkim.
LWT-Food Sci. Technol.61, 263-268.


FAO/WHO (2002) Guidelines for the Evaluation of Probiotics in Food. Report of a joint FAO/WHO Working Group on Drafting Guidelines for the Evaluation of Probiotics in Food. London, Ontario, Canada.

Hwang JH, Song YO, and Cheigh HS (2000) Fermentation characteristics and antioxidative effect of red mustard leaf kimchi.
J Korean Soc. Food Sci. Nutr.29, 1009-1015.

Kang JH, Yun SI, Park MH, Park JH, Jeong SY, and Park HO (2013) Anti-obesity effect of
Lactobacillus gasseri BNR17 in high-sucrose diet-induced obese mice.
PLoS ONE:
8, e54617.




Kim JY, Woo HJ, Kim YS, and Lee HJ (2002) Screening for antiproliferative effects of cellular components from lactic acid bacteria against human cancer cell lines.
Biotechnol. Lett.24, 1431-1436.


Kimura M (1983) The Neutral Theory of Molecular Evolution. Cambridge, Cambridge University Press.


Kluge AG and Farris JS (1969) Quantitative phyletics and the evolution of anurans.
Syst. Biol.18, 1-32.


Kumar S, Stecher G, and Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets.
Mol. Biol. Evol.33, 1870-1874.



Lane DJ (1991) 16S/23S rRNA sequencing. In: Nucleic Acid Techniques in Bacterial Systematics. E. Stackbrandt and M. Goodfellow(eds), John Wiley & Sons, Chichester, pp. 115-175.

Lin MY and Chang FY (2000) Antioxidative effect of intestinal bacteria
Bifidobacterium longum ATCC15708 and
Lactobacillus acidophilus ATCC 4356.
Dig. Dis. Sci.45, 1617-1622.

Oxman T, Shapira M, Diver A, Klein R, Avazov N and Rabinowitz B (2000) A new method of long-term preventive cardioprotection using
Lactobacillus.
Am. J. Physiol.278, 1717-1724.



Pieniz S, Andreazza R, Anghinoni T, Camargo F, and Brandelli A (2014) Probiotic potential, antimicrobial and antioxidant activities of
Enterococcus durans strain LAB18s.
Food Control.37, 251-256.


Rivera-Espinoza Y and Gallardo-Navarro Y (2010) Non-dairy probiotic products.
Food Microbiol.27, 1-11.



Roh SW, Sung Y, Nam YD, Chang HW, Kim KH, Yoon JH, Jeon CO, Oh HM, and Bae JW (2008)
Arthrobacter soli sp. nov., a novel bacterium isolated from wastewater reservoir sediment.
J. Microbiol.46, 40-44.



Saitou N and Nei M (1987) The neighbour-joining method: A new method for reconstructing phylogenetic trees.
Mol. Biol. Evol.4, 406-425.

Sánchez B, Champomier-Vergès MC, Collado MC, Anglade P, Baraige F, Sanz Y, delosReyes-Gavilán CG, Margolles A, Zagorec M (2007) Low-pH adaptation and the acid tolerance response of
Bifidobacterium longum biotypelongum.
Appl. Environ. Microbiol.73, 6450-6459.




Shin KS, Chae OW, Park IC, Hong SK, and Choe TB (1998) Antitumor effects of mice fed with cell lysate of
Lactobacillus plantarum isolated from kimchi.
Korean J. Biotechnol. Bioeng.13, 357-363.

Simone P, Robson A., Thiago A, Flávio C, and Adriano B (2014) Probiotic potential, antimicrobial and antioxidant activities of
Enterococcus durans strain LAB18s.
Food Control.37, 251-256.


Singh TP, Kaur G, Malik RK, Schillinger U, Guigas C, Kapila S (2012) Characterization of intestinal
Lactobacillus reuteri Strains as potential probiotics.
Probiotics & Antimicro. Prot.4, 47-58.



Spyropoulos BG, Misiakos EP, Fotiadis C, Stoidis CN (2011) Antioxidant properties of probiotics and their protective effects in the pathogenesis of radiation-induced enteritis and colitis.
Dig. Dis. Sci.56, 285-294.



Tulika A, Satvinder S, and Raj KS (2013) Probiotics: Interaction with gut microbiome and antiobesity potential.
Nutrition.29, 591-596.



Yadav H, Jain S, and Sinha PR (2007) Antidiabetic effect of probiotic dahi containing
Lactobacillus acidophilus and
Lactobacillus casei in high fructose fed rats.
Nutrition.23, 62-68.