INTRODUCTION
Lactic acid bacteria (LAB) have been recognized as part of the main fermentative agent in food fermentation with the Lactobacillus general being the most important in food processing within the group (Olasupo et al., 1997; Pasolli et al., 2020). For functional food development, LAB include the best microorganism used and they are abundant in nature especially in fermented food all over the world (Tamang and Samuel, 2010; Franz et al., 2014). Some traditional nourishments in Africa that contain LAB naturally are highlighted by Olasupo et al. (1997) and Franz et al. (2014) includes “Ogi”, “Fufu”, “Kunnu”, “Iru”, “Lafun”, “Ogiri”, “Nunu”, “Kindirmo” and “Wara” amidst many others. Most of these traditional foods are being consumed for ages, still needs optimization before they can be suitable to serve as a delivery vehicle for probiotics. However, as the important first step is to investigate the diversity of LAB found and associated with traditional food, this study has focused on kunun-zaki and kindirmo. Kunun-zaki is a cereal fermented drink (Gaffa et al., 2002) and the production involves soaking the millet for a period of time, grinding followed by sedimentation and afterwards a quarter is permitted to ferment at ambient temperature while the remaining is boiled before mixing the two parts together (Oyewole, 1997, Oguntoyinbo et al., 2012). At the time of production, naturally occurring LAB ferments the cereal which gave kunun-zaki its characteristic flavour and taste (Oguntoyinbo et al., 2012). While, Kindirmo a regular fermented milk amidst the Fulani herdsmen in Nigeria, despite of their nomadic nature it has been introduced to the mainstream populace (Sudi et al., 2011). It is produced by boiling raw milk and letting it to ferment naturally overnight. Often, a good portion of the former produce is kept for the inoculation of a new one serving as starter culture. The organoleptic property might vary from vendor to vendor due to difference in the type of starter culture used (Igwe et al., 2014), contaminant or change in seasonal temperature that might affect the rate of fermentation (Abdullahi et al., 2001).
These two traditional beverages are non-carbonated and non-alcoholic drink (Gaffa, 2002) from non-dairy and dairy sources widely accepted and consumed in various part of Nigeria irrespective of social class (Fapohunda and Adeware, 2012, Jaiyeoba et al., 2019). Although, Kunun-zaki (fermented millet—a non-dairy drink) is readily available and progressively being found across all states in Nigeria (Franz et al., 2014) whereas, Kindirmo (a dairy fermented drink) (Igwe et al., 2014) is consumed predominantly in the North (Abdullahi et al., 2001). Both are a product of chanced fermentation that involves diverse community of microorganism playing different roles in the process of fermentation (Gaffa and Gaffa, 2004).
Fermented foods traditionally produced have been consumed for many centuries across diverse culture depending on the cuisine of the geographical location (Heinen et al., 2020). The various fermented food from Africa have also been well documented and the continent is known to have diverse fermented food products (Oyewole, 1997; Holzapfel, 2002; Anukam and Reid, 2009, Tamang and Samuel, 2010). As a result of the active metabolic compound released by LAB that affects the sensory properties of food and their antimicrobial potential they are more desirous in the fermentation process as they elongate product’s shell life (Bamgbose, 2014; Dimidi et al., 2019; Heinen et al., 2020).
MATERIALS AND METHODS
Eighteen samples of kunun-zaki and thirteen samples of kindirmo were bought from local sellers from different outlets in Nigeria. They were kept in a sterile bottle and taken to the laboratory in ice pack for analysis. The samples were immediately prepared for isolation while the remaining were stored at 4°C and analysed within 48 h (Oguntoyinbo et al., 2012).
Samples were serially diluted using maximum recovery diluent (MRD) (Reuben et al., 2019). One millilitre aliquots of the samples were transferred into 9 ml MRD and serially diluted up to 10-9 dilutions. Each dilution was plated on selective medium for LAB de Man, Rogosa Sharpe (MRS) agar (Hi-Media, pH 6.4) (De Man et al., 1960) and incubated anaerobically at 37°C for 48 h following the procedure of Hawaz (2014). After incubation, single distinct colonies were chosen and further purified by streak plate technique and the individual colonies after incubation were inoculated into sterile MRS broth (Hi-Media, pH 6.5) (Bassyouni et al., 2012, Ida et al., 2017). All media used were prepared fresh every day and autoclaved at 121°C for 15 min.
The distinct colonies were subjected to Gram’s reaction and catalase test for presumptive selection of LAB (Hawaz 2014). The gram positive and catalase negative isolates were further characterized based on physiological and biochemical tests (Briugs, 1953; Bayili et al., 2019). Following the procedure of Kavitha and Devasena (2013), a thin smear was made on glass slide using colony of an overnight culture. The smear was then air dried, heat fixed and stained with crystal violet for 60 s and rinsed with water. Subsequently, Lugol’s iodine (mordant) was used to fix the primary stain and rinsed with water. Later, the slide was decolourised by acetone and rinsed with water. Safranin was used as counter stain and left for 30 s, which was afterward washed with distil water. The stained smear was air dried and observed under the light microscope (Olympus oic) using ×100 oil immersion objective. Further, 3% hydrogen peroxide was added to 1 mL of 18 h old cultures. The isolates, that did not produce gas bubbles, were selected as being catalase negative (Bayili et al., 2019).
Isolates that were identified as Gram positive and catalase negative were stored in MRS broth medium containing 20% (v/v) glycerol as frozen stocks at -20°C and -80°C. The glycerol stocks of samples were prepared by inoculating a loop full of overnight culture in a mixture of 1.2 mL MRS broth and 0.3 mL of 80% sterile glycerol in 2 mL cryovials following the procedure of Ida et al. (2017). To revive the isolate, a loop full of isolate from cryovial was streaked on the agar and incubated under anaerobic condition for 24 h – 48 h. Individual colony was picked and inoculated into another MRS broth medium and stored in -80°C freezer while other colony was used for further study.
The propagation of LAB isolates were studied in MRS broth at temperature 15°C and 45°C (Harrigan, 1998; El Soda et al., 2003) while growth at different sodium chloride (NaCl) concentrations and CO2 production from glucose (Hawaz, 2014) were carried out.
To test for the ability of isolates to grow at different temperature, MRS broth containing Bromothymol blue indicator was used as the test media and 5 mL of the medium transferred into test tubes. Afterwards, 50 μL of overnight culture was inoculated into the tubes and incubated for 10 days at 15°C and 7 days at 45°C (El Soda et al., 2003). During the incubation time, cell growth at each temperature was determined by the change of colour in the culture medium, from blue to yellow.
In order to see the ability of the isolate to tolerate lower pH, MRS broth was adjusted to pH 3.0 and overnight culture was inoculated into it and incubated at 37°C for 24 h (Bassyouni et al., 2012). 200 μL of suspended cell serves as positive control, 200 μL of broth with no suspended cell was used for monitoring contamination while 200 μL of MRS broth (pH 3) with no suspended cells served as negative control. Optical density was taken before and after incubation at OD = 600 nm to determine the LAB growth.
For further physiological differentiation, isolates were evaluated for their tolerance against different sodium chloride (NaCl) concentrations (2%, 4% and 6.5% w/v) (Shehata et al., 2016). Using a 96 well microtiter plate, 150 μL of NaCl medium were inoculated with 50 μL of overnight suspended cell cultures and then incubated at 37°C for 24 h (Kavitha and Devasena, 2013). 200 μL of suspended cell serves as positive control, 200 μL of broth with no suspended cell was used for monitoring contamination while 150 μL of medium plus 50 μL of broth with no suspended cells served as negative control 1 and 200 μL of NaCl medium served as negative control 2. Optical density was taken before and after incubation at OD = 600nm to determine the LAB growth.
To study the carbohydrate fermentation and other biochemical profile of each isolates, HiCarbohydrateTM kit (Cat# KB009, HiMedia) was used. Overnight culture of the isolate was centrifuged at 5,000 rpm for 10 min to harvest the cells; the cells were re-suspended in PBS (pH 7.4) and centrifuged at 5,000 rpm to wash it. Subsequently, 50 μL of the cells were adjusted to OD 0.5 at 600nm and inoculated on to the surface of the HiCarboTM wells using a sterile pipette. The strips were then incubated at 37°C for 24 h and the change in colour was observed and noted as positive reaction (+) or negative reaction (-) according to the colour change. A change from pinkish red to yellow indicates positive reactions for the carbohydrate as indicated in the manufacturer’s manual.
To know the fermentative pathway of each isolate, whether they are homofermentative or heterofermentative, CO2 production from glucose were tested for by using citrate lacking MRS broths using inverted Durham tubes in 1% overnight fresh cultures at 37°C for 24 h by following the method of Kavitha and Devasena (2013). Presence of gas raise up the Durham tubes is an evidence of CO2 production from glucose and thus a hetero-fermentative pathway (Briugs, 1953).
One millilitre (108cfu/mL) of actively grown culture of the isolates was pipetted into petri plates, followed by pouring 10 mL of MRS agar into it. The mixture was gently swirled, allowed to stand till it solidifies. With the aide of sterilized forceps, antibiotic discs (HiMedia, Mumbai) - erythromycin (10g), gentamycin (50g), vancomycin (10g) were aseptically positioned on the surface of the already solidified agar and kept in biosafety cabinet for 30 min for the antibiotics to diffuse. Afterwards, the petri plates were further incubated anaerobically at 37°C for 24 h according to the method of Ida et al. (2017). Based on clusters of different physiological and biochemical properties, thirty-four (34) isolates were selected for 16S rRNA molecular analysis.
The pure colony isolated was grown in 5 mL MRS broth at 37°C for 24 h in an anaerobic condition and was used for the extraction of the genomic DNA (gDNA) of the LAB. Further, broth culture was centrifuged (8,000 rpm, 10 min, 4°C) to harvest the cell. The harvested cells were washed in 1X PBS (pH 7.2) by centrifuging the cells at 8,000 rpm. The supernatant was removed and step repeated twice. Pellets were used for the DNA extraction by re-suspending it into 200 μL of 1X PBS. The DNA extraction were carried out using QIAampR DNA isolation kit (Qiagen, Germany, Lot:160052636) as per manufacture’s instruction. The DNA was eluted in 150 μL of elution buffer (provided with the kit) and the weight measured using NanoDrop spectrophotometer.
Extracted DNA was subjected to PCR using universal 27F and 1429R primers and specific primers for L. fermentum (Table 1) to amplify the 16s rRNA in the bacterial genome. The PCR reaction mixture consists of 12.5 μL of GoTaq® DNA polymerase (Promega; #cat.no. M7123); 1 μL (0.5 μM) of forward and reverse primer (GCC Biotech); 8.5 μL of nuclease free water and 2 μL of DNA template to make a final volume of 25 μL. The thermal cycler program was set as initial denaturation 95°C for 3 min, denaturation 94°C for 30 s, Annealing 52°C for 45 s, Extension 72°C for 1 min and final extension at 72°C for 10 min for 35 cycles carried out in Applied Biosystem (ABS).
| S/N | Primer | Sequence (5’→ 3’) |
|---|---|---|
| 1 | 27F | AGAGTTTGATCCTGGCTCAG |
| 1429R | GGTTACCTTGTTACGACTT | |
| 2 | Fermenticum-F | GACCAGCGCACCAAGTGATA |
| Fermenticum-R | AGCCTAGCGTTCGTGGTAAT |
The amplified PCR products were separated on 1% (w/v) agarose gel electrophoresis (Genetix Biotech Asia Pvt. Ltd.); band pattern was visualized by ethidium bromide (EtBr 0.5 mg/mL) and photographed in gel documentation system (Uvitech Cambridge).
All the amplified products were subjected to Sanger sequencing (Eurofins Genomics, Bengaluru, India) and the obtained nucleotide sequences were Nucleotide-BLAST in NCBI database. Similarity ≥ 97% with existing gene in NCBI database were used for confirmation of identity.
The evolutionary history was inferred using the Neighbor-Joining method. The optimal tree with the sum of branch length = 0.57724294 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) were shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the p-distance method and were in the units of the number of base differences per site. All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in MEGA6.
RESULTS
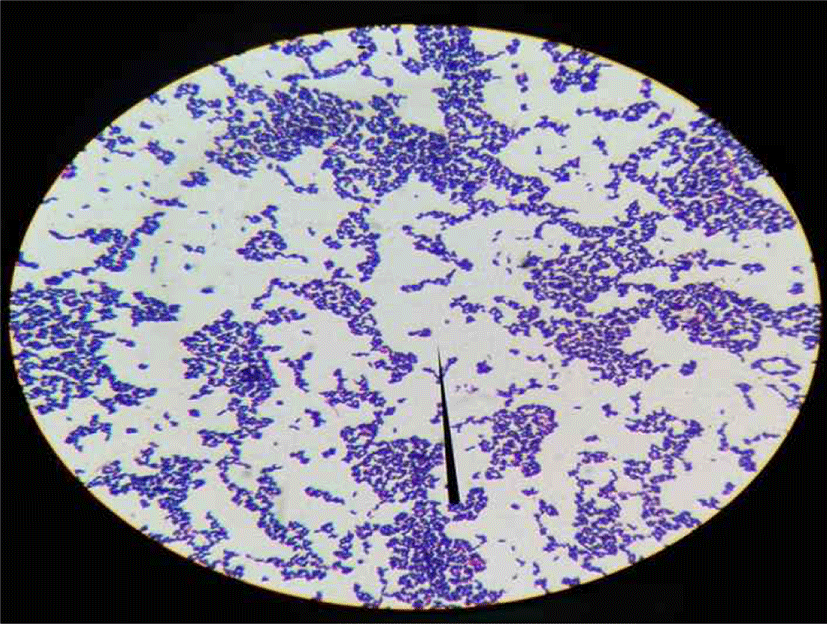
The population of isolated bacteria ranges from 105 – 107 for kunun-zaki and 108 - 109 for kindirmo. From the thirty one samples, eighty (80) isolates [66 kunun-zaki, 14 kindirmo] were presumptively selected as LAB based on their grams reaction, catalase test (Harrigan, 1998) and morphology (Fig. 1) (Sharpe, 1979). They were all Grams’ positive (Fig. 2) and catalase negative with varying morphology - bacilli (93.7%) and spherical (6.3%).
After 24 h of incubation at 37°C all isolates grew with a pattern also observed by Reuben et al. (2019) during the selection of LAB for poultry probiotic a credence to their optimum temperature. In order to differentiate further, LAB growth at 15°C and 45°C were investigated and divided into four groups based on growth pattern. Those that grew only at optimum temperature and could not tolerate 15°C and 45°C makes the highest number of the isolates at 55%, followed by 27.5% that grew only at 45°C. Only one isolate (1.25%) grew at both temperatures while 15% of the isolate tolerated 15°C but not 45°C. According to their thermo-tolerance, the probable organism (Table 2) is in-line with Briugs (1953) observation as the range of temperature LAB can tolerate determines its industrial application. However, there are many overlaps and usage of growth temperature for differential test is not accurate.
From the analysis, 59 (73.8%) isolates tolerated the acidic medium while 21 (26.2%) lost their viability (Table 3). The possibility that majority tolerated pH 3 above 3 h can be predicted and it’s in agreement with the reports of Argyri et al. (2013) and Reuben et al. (2019) of the ability of LABs to tolerate lower pH.
The growth pattern to different salt concentration alongside other physiological and biochemical properties is in accordance to the description in Bergey’s manual of determinative bacteriology similar to reports by Shehata et al. (2016) in screening for LAB for lowering cholesterol. All isolates have distinctive pattern of salt toleration (Table 4) which is in variance to the report of Hawaz, (2014) where all isolated LABs tolerated 2%, 4% and 6.5% salt concentration.
| Group | NaCl concentration | Count (%) (n=80) | ||
|---|---|---|---|---|
| 2% | 4% | 6.5% | ||
| I | + | + | + | 26 (32.5) |
| II | + | + | - | 21 (26.25) |
| III | + | - | - | 22 (27.5) |
| IV | + | - | + | 0 (0) |
| V | - | + | - | 3 (3.75) |
| VI | - | + | + | 2 (2.5) |
| VII | - | - | + | 1 (1.25) |
| VIII | - | - | - | 5 (6.25) |
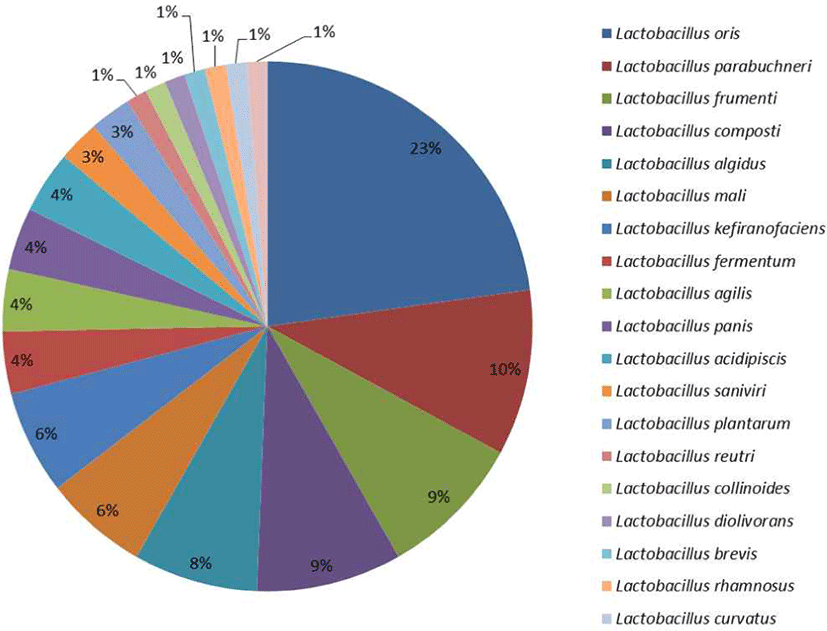
All the LAB isolates were able to ferment glucose to produce lactic acid, although, as noted by Briugs (1953) the carbohydrate fermentation profile is not a definitive means for classification as it is strain dependent and gives a varying result depending on other factors of growth. Thus, it is problematic to separate the isolates especially the closely related ones based on their fermentation profile. However, it gave an understanding and a lead on their bacteriological classification (Dowarah et al., 2018). Lactobacillus oris were the most probable organism according to the fermentation profile (Supplementary 4) followed by Lactobacillus fermentum which was later confirmed by molecular method. Lactobacillus rhamnosus, L. curvatus, L. brevis were the list predominant (Fig. 3).
From this study, 95% of the isolates were resistant to vancomycin, while 46.8% were resistant to gentamycin with erythromycin recording the lowest at 3.2% resistance (Supplementary 6 ). Lactic acid bacteria in many reports have shown to be susceptible to erythromycin while they have natural resistance ability to vancomycin (Fguiri et al., 2016).
From the thirty-four isolates subjected to molecular identification using universal primers, three genera [Limosilactobacillus (68%); Lactiplantibacillus (6%) and Weissella (3%)] were identified while Pediococcus (3%); Oenococcus (3%); Lactococcus (3%) and Leuconostoc (3%) had a percentage similarity less than 97% and their identity was not ascertained (Table 5). Considering that Limosilactobacillus was dominant, specific primer for L. fermentum was used to amplify DNA of a further thirty-two isolates with fourteen (14) of the isolates confirmed as L. fermentum. Limosilactobacillus fermentum was the most common LAB associated to both food samples, followed by Lactiplantibacillus plantarum (Table 5). The molecular relationship between the isolates is illustrated by a phylogenetic tree (Fig. 4).

DISCUSSION
In traditional fermented foods, LAB considerably are crucial for the overall fermentation process (Aspri et al., 2020), product flavor (Dinesh, 2015; Bayili et al., 2019), taste (Adaramola-Ajibola et al., 2019) and shelf life (Ikpoh et al., 2013; Silva et al. 2018). The local producer have virtually no knowledge of the role of microorganism behind the process; but in order to scale up the production, the common LABs involved in the fermentation need to be identified and characterized for commercial use as starter culture, nutrient improvement, safety purpose and development into a probiotic product. Probiotic market in Africa is still very limited, hence there is need to have more investigation on the diversity of LAB present in the traditionally fermented food; one in which this study have focused on identifying while further study on their probiotic potential is essential.
From the study there was no statistically significant difference between the isolates that grew and did not grow at 15°C (F(2,77) = 0.151, (p = 0.070) or at 45°C (F(2,77) = 0.391, (p = 0.069) while there was a statistically significant difference between their ability to grow at pH 3 and their fermentation pathway (F(2,77) = 4.267, (p = 0.017). This reveals that the fermentation pathway has no correlation with the temperature of growth while it is unlikely due to chance that their ability to grow at pH 3 is dependent on their fermentation pathway. The ability for the isolate to grow in an acidic condition is an important selection criterion for LAB (Sharpe, 1979; Teuber, 2008; Reuben et al., 2019) and in the present study 73.8% of the strains were able to tolerate a lower pH indicating that 26.2% of the isolate are not desirable for acidic industrial application (Table 3). The strains that grow at higher temperature of 45 °C and are able to tolerate pH 3 stood at 33.9%, significantly this isolate can be explored for industrial production that involves high heat (Luo et al., 2011; Amarantini et al., 2019) while 23.9% of the strains that makes up the isolates that grows at 15°C can be useful in improving technological properties and shelf-life of food product that needs cold storage for stability (Mombelli and Gismondo, 2000; Kisan et al., 2019).
Likewise, as part of the physiological property is the growth pattern to different salt concentration alongside other physiological and biochemical properties. High salt can damage the activities of certain enzymes thereby affecting bacterial physiology and metabolism (Olajugbagbe et al., 2020). Each strain has a different composition of membrane phospholipid and the ATP-dependent glycine is strain-specific too, thus the tolerance to osmotic pressure exerted by high salt varies with each isolate a pattern that was noticeable in this study (Table 4). Hence, the pattern observed in this study is in accordance with the description reported by Shehata et al. (2016) in screening for LAB for lowering cholesterol. Nevertheless, the result is different to the report of Hawaz (2014) where all isolated LABs tolerated 2%, 4% and 6.5% salt concentration. Prabhurajeshwar and Chandrakanth (2019) reported in their study that Lactobacillus sp. isolated from yoghurt tolerates NaCl concentration up to 9 % (w/v) but the best growth is observed between 1 % - 5 % (w/v) NaCl concentrations.
The carbohydrate profile of the LAB isolates was studied and all showed the ability to ferment glucose to produce lactic acid. Relying on carbohydrate fermentation profile for identification can be misleading, as it is usually not accurate enough to identify up to species level as several LAB species have similar physiological characteristics (Dowarah et al. 2018). In this study, the identification to genus level by carbohydrate fermentation profile is accurate when compared to the molecular result while only 4 % of the isolate were accurately identified up to species level. The result is similar to Dowarah et al. (2018) where all LAB isolates were able to utilize maltose, fructose, dextrose and mannose and the majority showed a positive reaction for the fermentation of lactose, galactose, trehalose, and mellobiose. Although, the sources of the LAB isolates varied widely in their study, while this study the LAB isolates were from fermented drinks. The similarity in fermentation profile will give misinformation on their relatedness if considered solely. As noted by Reuben et al. (2019) the identification of LAB by carbohydrate profile is very inconsistent and should not be relied on for species-level identification. Thus, it is problematic to separate the isolates especially the closely related ones based on their fermentation profile.
One important benefit for the ability of the isolates to utilize lactose is their possible usage in the treatment of lactose-intolerant individuals. Dimidi et al. (2019) reported that keffir – a fermented milk is the most investigated fermented food for lactose-intolerance treatment as lactose-fermenting LAB have been isolated from it and studied while Valero-cases et al. (2020) have suggested that non-dairy fermented beverages can be the solution for people that are lactose-intolerant and cannot consume the dairy product. In this present study, the focus has been on both dairy and non-dairy fermented products with 95% of the isolate having the ability to utilize lactose (Supplementary 4). Hence, kunun-zaki can be developed as a non-dairy carrier for lactose-fermenting probiotics for the management of lactose-intolerant individuals. Such strain can be used as well for the control or industrial fermentation of kindirmo that will have the property of reducing the lactose concentration by hydrolysing the lactose in the drink thereby allowing people suffering from lactose mal-absorption to tolerate the consumption of kindirmo.
In addition, considering that antibiotic resistance is a major food safety concern in production, it will be undesirable to use LAB that have the propensity to transfer antibiotic resistance gene to other organisms (Vieco-Saiz et al., 2019). Vancomycin is an example of an antimicrobial agent that LAB has intrinsic resistance. Vancomycin sensitivity has also been utilised in the delineation of LAB in bacteria taxonomy (Zheng et al., 2020). Thus, resistance to vancomycin (Zhang et al., 2016) and in some cases gentamicin (Makete, 2016) is one of the crucial indicators in LAB antibiotic resistant pattern for selection for further study as a potential probiotic as they do not carry the resistant gene in the plasmid but in the chromosome, which is not transferrable by horizontal gene transfer. The antibiotic resistant pattern was studied in order to determine whether a LAB isolate is resistant to antibiotics it should be naturally susceptible too and in the case of resistance chances are it has been acquired; such strain will not be suitable for further use according to CLSI guidelines. From this study 10% of the isolate had mild sensitivity between 10mm to 25mm to vancomycin (10 μg). As resistance to vancomycin in LAB is intrinsic, those that showed some forms of susceptibility were not considered as a suitable candidate for further study. The result from this study is in agreement with Makete (2016) where all LAB isolated from milk of South African Saanen goats were all resistant to vancomycin and gentamicin. In addition, Zhang (2016) reported 100% resistance to vancomycin and streptomycin by LAB isolated from traditional Tibetan qula – a raw yak milk. This study is also similar to the report of Shehata et al. (2020) that described eight different LAB isolated from traditionally fermented milk in Egypt that has 100% resistance to vancomycin.
Taking into consideration all the physiological and biochemical factors, 34 strains were selected for 16S rRNA gene sequence analysis that identified 3 genera of the LAB with Limosilactobacillus being the predominant genera (Table 5). The genus Lactobacillus with over 255 species has been reclassified into 25 different genera and the original genus Lactobacillus that was described from Lactobacillus delbrueckii in 1901 retaining only 35 species while the remaining were placed under Paralactobacillus and 23 other genera (Claesson et al., 2008; Salvetti et al., 2018; Zheng et al., 2020). The genera Limosilactobacillus makes up the highest number of LAB isolated in line with reports of Lactobacillus fermentum from Kunun-zaki by Franz et al. (2014) and Lactobacillus plantarum from kindirmo (Igwe et al., 2014) which is similar to the result in this study. The report from this study is also comparable to those of Olasupo et al. (1997), Todorov et al. (2008) and Fapohunda and Adeware, (2012). In addition, the isolates from this study (Fig. 3) has a close relationship with other species from different sources and location. This indicates that though they were isolated from separate sources their genetic relationship does not differ much especially in terms of taxonomy. However, if complete genome was carried out there will be quite a few specific genes that distinguishes them at strain level.
The hallmark of this study was to identify the presence of LAB (LAB) involved in the chance fermentation of the two traditionally fermented food samples. Combining the physiological and biochemical properties for the identification of LAB and confirming the identity of isolates with molecular tool, the presence of LAB were established. Also, the isolates were grouped to 16 clusters based on their morphological identity and the sugar utilization profile across the group varied widely indicating variations at the strain level (Dowarah et al., 2018).
Moreover, the ability of each isolates to tolerate diverse range of temperature, salinity and carbohydrate fermentation pathway points to the huge diversity at the species level of the isolates which present a huge advantage for more study of all individual organism for their specific properties. Consequently, from the antibiotics sensitivity pattern, none of the isolates appears to carry transferrable antibiotic resistant gene as they were resistant and susceptible to antibiotics in same pattern as highlighted in CLSI guidelines and in line with most research submission making them a good candidate for industrial process (Zhang et al., 2016; Fguiri et al., 2016; Bayili et al., 2019). To the best of our knowledge this is the first study that focused majorly on characterizing the LAB present in kunun-zaki and kindirmo for the purpose of identifying the predominant LAB that can be studied further for their probiotic potential. Majority of the study focuses on sensory characteristics (Adaramola-Ajibola et al., 2019; Jaiyeoba et al., 2019), variations in the starter culture (Igwe et al., 2014), microbiological quality and bacteria succession (Abdullahi et al., 2001; Gaffa and Gaffa, 2004; Fapohunda and Adeware, 2012) in kunun-zaki and kindirmo.
CONCLUSION
The combination of physiological and biochemical tests have been used to characterise LAB isolated while 16S rRNA gene have been used for the confirmation of their identity. Each strain has its spectrum of temperature, acidity and salinity it can tolerate while the carbohydrate fermentation pathways are divergent and strain specific. Moreover, LAB isolated from this study are mostly from Limosilactobacillus genera which are in line with couple of studies on other fermented drink from West Africa. It is to be noted that Lactobacillus is one of the major microorganisms used for probiotic development (Heinen et al., 2020; Pasolli et al., 2020) and this study further cement the concept that traditionally fermented foods are a good natural source for beneficial bacteria. Hence, some of the strains can be investigated further in order to determine their suitability as a starter culture for the industrial production of kunun-zaki and kindirmo. Additionally, study on their ability to produce bacteriocin for application in food preservation and probiotic potential for functional food development in medical application is a good future prospect worth exploring in addition to their probiotic potential.







